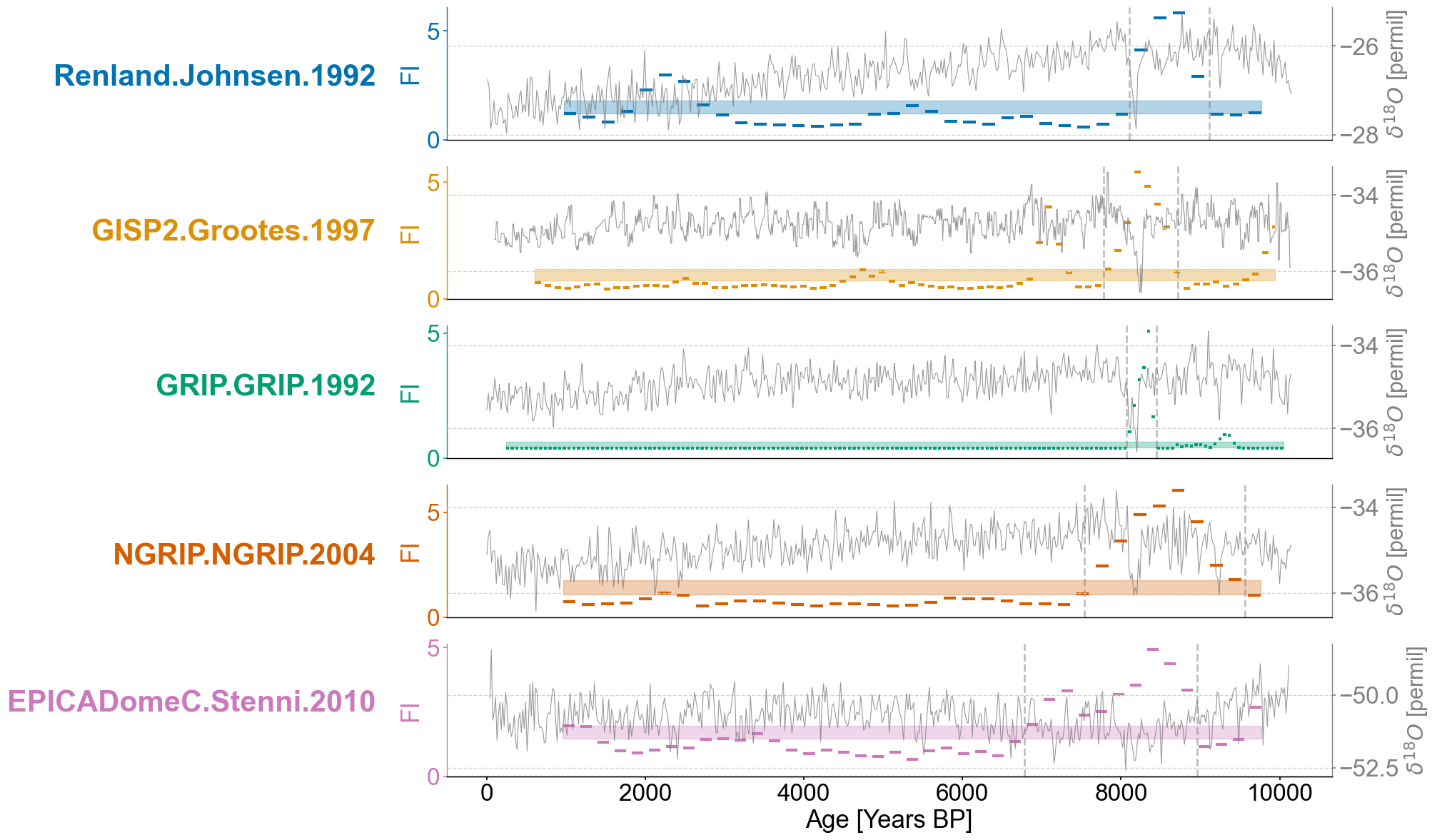
Figure 5#
import pickle
import random
import pyleoclim as pyleo
import matplotlib.pyplot as plt
from matplotlib import gridspec
import matplotlib.transforms as transforms
import matplotlib.patches as mpatches
import seaborn as sns
import numpy as np
import ammonyte as amt
from tqdm import tqdm
from pylipd.lipd import LiPD
#We suppress warnings for these notebooks for presentation purposes. Best practice is to not do this though.
import warnings
warnings.filterwarnings('ignore')
Analysis here is largely identical to that for Figures 2 and 3, though we use slightly different parameters.
def detect_transitions(series,transition_interval=None):
'''Function to detect transitions across a confidence interval
Parameters
----------
series : pyleo.Series, amt.Series
Series to detect transitions upon
transition_interval : list,tuple
Upper and lower bound for the transition interval
Returns
-------
transitions : list
Timing of the transitions of the series across its confidence interval
'''
series_fine = series.interp(step=1)
if transition_interval is None:
upper, lower = amt.utils.sampling.confidence_interval(series)
else:
upper, lower = transition_interval
above_thresh = np.where(series_fine.value > upper,1,0)
below_thresh = np.where(series_fine.value < lower,1,0)
transition_above = np.diff(above_thresh)
transition_below = np.diff(below_thresh)
upper_trans = series_fine.time[1:][np.diff(above_thresh) != 0]
lower_trans = series_fine.time[1:][np.diff(below_thresh) != 0]
full_trans = np.zeros(len(transition_above))
last_above = 0
last_below = 0
for i in range(len(transition_above)):
above = transition_above[i]
below = transition_below[i]
if above != 0:
if last_below+above == 0:
loc = int((i+below_pointer)/2)
full_trans[loc] = 1
last_below=0
last_above = above
above_pointer = i
if below != 0:
if last_above + below == 0:
loc = int((i+above_pointer)/2)
full_trans[loc] = 1
last_above=0
last_below = below
below_pointer = i
transitions = series_fine.time[1:][full_trans != 0]
return transitions
color_list = sns.color_palette('colorblind')
Here we load the data using pyLiPD.
lipd_path = './data/8k_ice'
all_files = LiPD()
if __name__=='__main__':
all_files.load_from_dir(lipd_path,parallel=True)
record_names = all_files.get_all_dataset_names()
Directory ./data/8k_ice does not exist
series_list = []
# We specify the indices of interest in each dataframe by hand here
index_dict = {
'GRIP.GRIP.1992' : 'd18O',
'Renland.Johnsen.1992' : 'd18O',
'EDML.Stenni.2010' : 'bagd18O',
'EPICADomeC.Stenni.2010' : 'bagd18O',
'Vostok.Vimeux.2002' : 'temperature',
'GISP2.Grootes.1997' : 'd18O',
'NGRIP.NGRIP.2004' : 'd18O',
'TALDICE.Mezgec.2017' : 'd18O',
}
for record in record_names:
d = LiPD()
d.load(f'{lipd_path}/{record}.lpd')
df = d.get_timeseries_essentials()
row = df[df['paleoData_variableName']==index_dict[record]][df['time_variableName']=='age']
lat = row['geo_meanLat'].to_numpy()[0]
lon = row['geo_meanLon'].to_numpy()[0]
elevation = row['geo_meanElev'].to_numpy()[0]
value = row['paleoData_values'].to_numpy()[0]
value_name = row['paleoData_variableName'].to_numpy()[0]
value_unit = row['paleoData_units'].to_numpy()[0]
time = row['time_values'].to_numpy()[0]
time_unit = row['time_units'].to_numpy()[0]
time_name = row['time_variableName'].to_numpy()[0]
label = row['dataSetName'].to_numpy()[0]
geo_series = pyleo.GeoSeries(time=time,
value=value,
lat=lat,
lon=lon,
elevation=elevation,
time_unit=time_unit,
time_name=time_name,
value_name=value_name,
value_unit=value_unit,
label=label,
archiveType='ice')
series_list.append(geo_series)
geo_ms = pyleo.MultipleGeoSeries(series_list)
greenland_ms_list = []
antarctica_ms_list = []
for series in geo_ms.series_list:
if series.lat > 0 or series.label == 'EPICADomeC.Stenni.2010':
series.time_unit = 'Years BP'
greenland_ms_list.append(series)
else:
series.time_unit = 'Years BP'
antarctica_ms_list.append(series)
end_time=10150
greenland_ms = pyleo.MultipleSeries([series.slice((0,end_time)).interp() for series in greenland_ms_list])
greenland_ms.stackplot(colors=color_list[:len(greenland_ms.series_list)],figsize=(8,10))
ms_dict = {series.label:series for series in greenland_ms.series_list}
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
Cell In[8], line 4
1 end_time=10150
3 greenland_ms = pyleo.MultipleSeries([series.slice((0,end_time)).interp() for series in greenland_ms_list])
----> 4 greenland_ms.stackplot(colors=color_list[:len(greenland_ms.series_list)],figsize=(8,10))
6 ms_dict = {series.label:series for series in greenland_ms.series_list}
File ~/miniconda3/envs/ammonyte/lib/python3.10/site-packages/pyleoclim/core/multipleseries.py:1866, in MultipleSeries.stackplot(self, figsize, savefig_settings, time_unit, xlim, fill_between_alpha, colors, cmap, norm, labels, ylabel_fontsize, spine_lw, grid_lw, label_x_loc, v_shift_factor, linewidth, plot_kwargs)
1863 raise ValueError("The length of the label list should match the number of timeseries to be plotted")
1865 # deal with time units
-> 1866 self = self.convert_time_unit(time_unit=time_unit)
1868 # Deal with plotting arguments
1869 if type(plot_kwargs)==dict:
File ~/miniconda3/envs/ammonyte/lib/python3.10/site-packages/pyleoclim/core/multipleseries.py:268, in MultipleSeries.convert_time_unit(self, time_unit)
266 for i, u in enumerate(unique_units):
267 count_units[i] = units.count(u)
--> 268 time_unit = unique_units[count_units.argmax()]
270 new_ms = self.copy()
271 new_ts_list = []
ValueError: attempt to get argmax of an empty sequence
lp_series_dict = {}
m = 12
tau = 4
for idx,series in enumerate(greenland_ms.series_list):
if series.label != 'EPICADomeC.Stenni.2010':
amt_series = amt.Series(
time=series.time,
value=series.value,
time_name = series.time_name,
value_name = series.value_name,
time_unit = series.time_unit,
value_unit = series.value_unit,
label = series.label,
clean_ts=False,
sort_ts=None
).convert_time_unit('Years').detrend(method='savitzky-golay')
else:
amt_series = amt.Series(
time=series.time,
value=series.value,
time_name = series.time_name,
value_name = series.value_name,
time_unit = series.time_unit,
value_unit = series.value_unit,
label = series.label,
clean_ts=False,
sort_ts=None
).convert_time_unit('Years')
td = amt_series.embed(m,tau)
print(f'{series.label} Tau is: {td.tau}')
eps = td.find_epsilon(eps=1,target_density=.05,tolerance=.01)
rm = eps['Output']
lp = rm.laplacian_eigenmaps(w_size=20,w_incre=4).convert_time_unit('Years BP')
lp_series_dict[series.label] = lp
/var/folders/5k/0y4jsz592qq0y78c_0ddgcpm0000gn/T/ipykernel_1021/3962517463.py:7: UserWarning: auto_time_params is not specified. Currently default behavior sets this to True. In a future release this will be changed to False.
amt_series = amt.Series(
/Users/alexjames/Documents/GitHub/Pyleoclim_util/pyleoclim/utils/tsutils.py:985: UserWarning: Timeseries is not evenly-spaced, interpolating...
warnings.warn("Timeseries is not evenly-spaced, interpolating...")
Renland.Johnsen.1992 Tau is: 4
Initial density is 0.0424
Initial density is within the tolerance window!
/Users/alexjames/Documents/GitHub/Ammonyte/ammonyte/core/rqa_res.py:22: UserWarning: auto_time_params is not specified. Currently default behavior sets this to True. In a future release this will be changed to False.
super().__init__(time,value,time_name,time_unit,value_name,value_unit,label,sort_ts=None)
/Users/alexjames/Documents/GitHub/Ammonyte/ammonyte/core/rqa_res.py:22: UserWarning: No time_name parameter provided. Assuming "Time".
super().__init__(time,value,time_name,time_unit,value_name,value_unit,label,sort_ts=None)
/var/folders/5k/0y4jsz592qq0y78c_0ddgcpm0000gn/T/ipykernel_1021/3962517463.py:7: UserWarning: auto_time_params is not specified. Currently default behavior sets this to True. In a future release this will be changed to False.
amt_series = amt.Series(
/Users/alexjames/Documents/GitHub/Pyleoclim_util/pyleoclim/utils/tsutils.py:985: UserWarning: Timeseries is not evenly-spaced, interpolating...
warnings.warn("Timeseries is not evenly-spaced, interpolating...")
NGRIP.NGRIP.2004 Tau is: 4
Initial density is 0.0217
Initial density is not within the tolerance window, searching...
Epsilon: 1.0000, Density: 0.0217
Epsilon: 1.1416, Density: 0.0583
Epsilon: 1.1416, Density: 0.0583.
/Users/alexjames/Documents/GitHub/Ammonyte/ammonyte/core/rqa_res.py:22: UserWarning: auto_time_params is not specified. Currently default behavior sets this to True. In a future release this will be changed to False.
super().__init__(time,value,time_name,time_unit,value_name,value_unit,label,sort_ts=None)
/Users/alexjames/Documents/GitHub/Ammonyte/ammonyte/core/rqa_res.py:22: UserWarning: No time_name parameter provided. Assuming "Time".
super().__init__(time,value,time_name,time_unit,value_name,value_unit,label,sort_ts=None)
/var/folders/5k/0y4jsz592qq0y78c_0ddgcpm0000gn/T/ipykernel_1021/3962517463.py:7: UserWarning: auto_time_params is not specified. Currently default behavior sets this to True. In a future release this will be changed to False.
amt_series = amt.Series(
/Users/alexjames/Documents/GitHub/Pyleoclim_util/pyleoclim/utils/tsutils.py:985: UserWarning: Timeseries is not evenly-spaced, interpolating...
warnings.warn("Timeseries is not evenly-spaced, interpolating...")
GISP2.Grootes.1997 Tau is: 4
Initial density is 0.0268
Initial density is not within the tolerance window, searching...
Epsilon: 1.0000, Density: 0.0268
Epsilon: 1.1158, Density: 0.0605
Epsilon: 1.1158, Density: 0.0605
Epsilon: 1.0631, Density: 0.0426
Epsilon: 1.0631, Density: 0.0426.
/Users/alexjames/Documents/GitHub/Ammonyte/ammonyte/core/rqa_res.py:22: UserWarning: auto_time_params is not specified. Currently default behavior sets this to True. In a future release this will be changed to False.
super().__init__(time,value,time_name,time_unit,value_name,value_unit,label,sort_ts=None)
/Users/alexjames/Documents/GitHub/Ammonyte/ammonyte/core/rqa_res.py:22: UserWarning: No time_name parameter provided. Assuming "Time".
super().__init__(time,value,time_name,time_unit,value_name,value_unit,label,sort_ts=None)
/var/folders/5k/0y4jsz592qq0y78c_0ddgcpm0000gn/T/ipykernel_1021/3962517463.py:19: UserWarning: auto_time_params is not specified. Currently default behavior sets this to True. In a future release this will be changed to False.
amt_series = amt.Series(
EPICADomeC.Stenni.2010 Tau is: 4
Initial density is 0.0022
Initial density is not within the tolerance window, searching...
Epsilon: 1.4777, Density: 0.0132
Epsilon: 1.8461, Density: 0.0653
Epsilon: 1.6928, Density: 0.0351
Epsilon: 1.8418, Density: 0.0641
Epsilon: 1.7009, Density: 0.0364
Epsilon: 1.8372, Density: 0.0631
Epsilon: 1.7065, Density: 0.0372
Epsilon: 1.8346, Density: 0.0625
Epsilon: 1.7095, Density: 0.0377
Epsilon: 1.8322, Density: 0.0619
Epsilon: 1.7137, Density: 0.0384
Epsilon: 1.8298, Density: 0.0613
Epsilon: 1.7171, Density: 0.0389
Epsilon: 1.8285, Density: 0.0610
Epsilon: 1.7188, Density: 0.0391
Epsilon: 1.8277, Density: 0.0609
Epsilon: 1.7190, Density: 0.0392
Epsilon: 1.8272, Density: 0.0608
Epsilon: 1.8272, Density: 0.0608
Epsilon: 1.7733, Density: 0.0491
Epsilon: 1.7733, Density: 0.0491.
/Users/alexjames/Documents/GitHub/Ammonyte/ammonyte/core/rqa_res.py:22: UserWarning: auto_time_params is not specified. Currently default behavior sets this to True. In a future release this will be changed to False.
super().__init__(time,value,time_name,time_unit,value_name,value_unit,label,sort_ts=None)
/Users/alexjames/Documents/GitHub/Ammonyte/ammonyte/core/rqa_res.py:22: UserWarning: No time_name parameter provided. Assuming "Time".
super().__init__(time,value,time_name,time_unit,value_name,value_unit,label,sort_ts=None)
/var/folders/5k/0y4jsz592qq0y78c_0ddgcpm0000gn/T/ipykernel_1021/3962517463.py:7: UserWarning: auto_time_params is not specified. Currently default behavior sets this to True. In a future release this will be changed to False.
amt_series = amt.Series(
/Users/alexjames/Documents/GitHub/Pyleoclim_util/pyleoclim/utils/tsutils.py:985: UserWarning: Timeseries is not evenly-spaced, interpolating...
warnings.warn("Timeseries is not evenly-spaced, interpolating...")
GRIP.GRIP.1992 Tau is: 4
Initial density is 0.1646
Initial density is not within the tolerance window, searching...
Epsilon: 0.0000, Density: 0.0000
Epsilon: 0.5000, Density: 0.0026
Epsilon: 0.5000, Density: 0.0026
Epsilon: 0.7371, Density: 0.0250
Epsilon: 0.7371, Density: 0.0250
Epsilon: 0.8622, Density: 0.0701
Epsilon: 0.8622, Density: 0.0701
Epsilon: 0.7617, Density: 0.0313
Epsilon: 0.7617, Density: 0.0313
Epsilon: 0.8554, Density: 0.0667
Epsilon: 0.8554, Density: 0.0667
Epsilon: 0.7719, Density: 0.0342
Epsilon: 0.7719, Density: 0.0342
Epsilon: 0.8511, Density: 0.0646
Epsilon: 0.8511, Density: 0.0646
Epsilon: 0.7781, Density: 0.0360
Epsilon: 0.7781, Density: 0.0360
Epsilon: 0.8479, Density: 0.0631
Epsilon: 0.8479, Density: 0.0631
Epsilon: 0.7824, Density: 0.0374
Epsilon: 0.7824, Density: 0.0374
Epsilon: 0.8454, Density: 0.0619
Epsilon: 0.8454, Density: 0.0619
Epsilon: 0.7860, Density: 0.0385
Epsilon: 0.7860, Density: 0.0385
Epsilon: 0.8432, Density: 0.0609
Epsilon: 0.8432, Density: 0.0609
Epsilon: 0.7886, Density: 0.0395
Epsilon: 0.7886, Density: 0.0395
Epsilon: 0.8414, Density: 0.0601
Epsilon: 0.8414, Density: 0.0601
Epsilon: 0.7909, Density: 0.0402
Epsilon: 0.7909, Density: 0.0402.
/Users/alexjames/Documents/GitHub/Ammonyte/ammonyte/core/rqa_res.py:22: UserWarning: auto_time_params is not specified. Currently default behavior sets this to True. In a future release this will be changed to False.
super().__init__(time,value,time_name,time_unit,value_name,value_unit,label,sort_ts=None)
/Users/alexjames/Documents/GitHub/Ammonyte/ammonyte/core/rqa_res.py:22: UserWarning: No time_name parameter provided. Assuming "Time".
super().__init__(time,value,time_name,time_unit,value_name,value_unit,label,sort_ts=None)
ms = greenland_ms
ordered_list = [
'Renland.Johnsen.1992',
'GISP2.Grootes.1997',
'GRIP.GRIP.1992',
'NGRIP.NGRIP.2004',
'EPICADomeC.Stenni.2010'
]
SMALL_SIZE = 22
MEDIUM_SIZE = 24
BIGGER_SIZE = 25
plt.rc('font', size=SMALL_SIZE) # controls default text sizes
plt.rc('axes', titlesize=SMALL_SIZE) # fontsize of the axes title
plt.rc('axes', labelsize=MEDIUM_SIZE) # fontsize of the x and y labels
plt.rc('xtick', labelsize=MEDIUM_SIZE) # fontsize of the tick labels
plt.rc('ytick', labelsize=MEDIUM_SIZE) # fontsize of the tick labels
plt.rc('legend', fontsize=SMALL_SIZE) # legend fontsize
plt.rc('figure', titlesize=BIGGER_SIZE) # fontsize of the figure title
fig,axes = plt.subplots(nrows=len(ordered_list),ncols=1,sharex=True,figsize=(16,14))
transition_timing = []
for idx,record in enumerate(ordered_list):
ts = lp_series_dict[record]
ts.value_name = 'FI'
ts.value_unit = None
ax = axes[idx]
ts_smooth = amt.utils.fisher.smooth_series(series=ts,block_size=3) #Using a block size of 3 for smoothing the Fisher information
upper, lower = amt.utils.sampling.confidence_interval(series=ts,upper=95,lower=5,w=50,n_samples=10000) #Calculating the bounds for our confidence interval using default values
transitions=detect_transitions(ts_smooth,transition_interval=(upper,lower))
transition_timing=[]
for transition in transitions:
if ts.label == "GISP2.Grootes.1997":
if transition > 7500 and transition < 9000:
transition_timing.append(transition)
elif ts.label == "GRIP.GRIP.1992":
if transition > 7500 and transition < 8700:
transition_timing.append(transition)
elif ts.label == "EPICADomeC.Stenni.2010":
if transition > 6500 and transition < 9400:
transition_timing.append(transition)
elif ts.label == "NGRIP.NGRIP.2004":
if transition > 6500:
transition_timing.append(transition)
else:
if transition > 6200 and transition < 10000:
transition_timing.append(transition)
ts.confidence_smooth_plot(
ax=ax,
background_series = ms_dict[record].slice((0,end_time)),
transition_interval=(upper,lower),
block_size=3,
color=color_list[idx],
figsize=(12,6),
legend=True,
lgd_kwargs={'loc':'upper left'},
hline_kwargs={'label':None},
background_kwargs={'ylabel':'$\delta^{18}O$ [permil]','legend':False,'linewidth':.8,'color':'grey','alpha':.8})
for transition in transition_timing:
ax.axvline(transition,color='grey',linestyle='dashed',alpha=.5)
trans = transforms.blended_transform_factory(ax.transAxes, ax.transData)
ax.text(x=-.08, y = 2.5, s = ts.label, horizontalalignment='right', transform=trans, color=color_list[idx], weight='bold',fontsize=30)
ax.spines['left'].set_visible(True)
ax.spines['right'].set_visible(False)
ax.yaxis.set_label_position('left')
ax.yaxis.tick_left()
ax.get_legend().remove()
ax.set_title(None)
ax.grid(visible=False,axis='y')
if idx != len(lp_series_dict.keys())-1:
ax.set_xlabel(None)
ax.spines[['bottom']].set_visible(False)
ax.tick_params(bottom=False)
ax.xaxis.label.set_fontsize(25)
ax.yaxis.label.set_fontsize(25)
ax.set_yticks(ticks=np.array([0,5]))