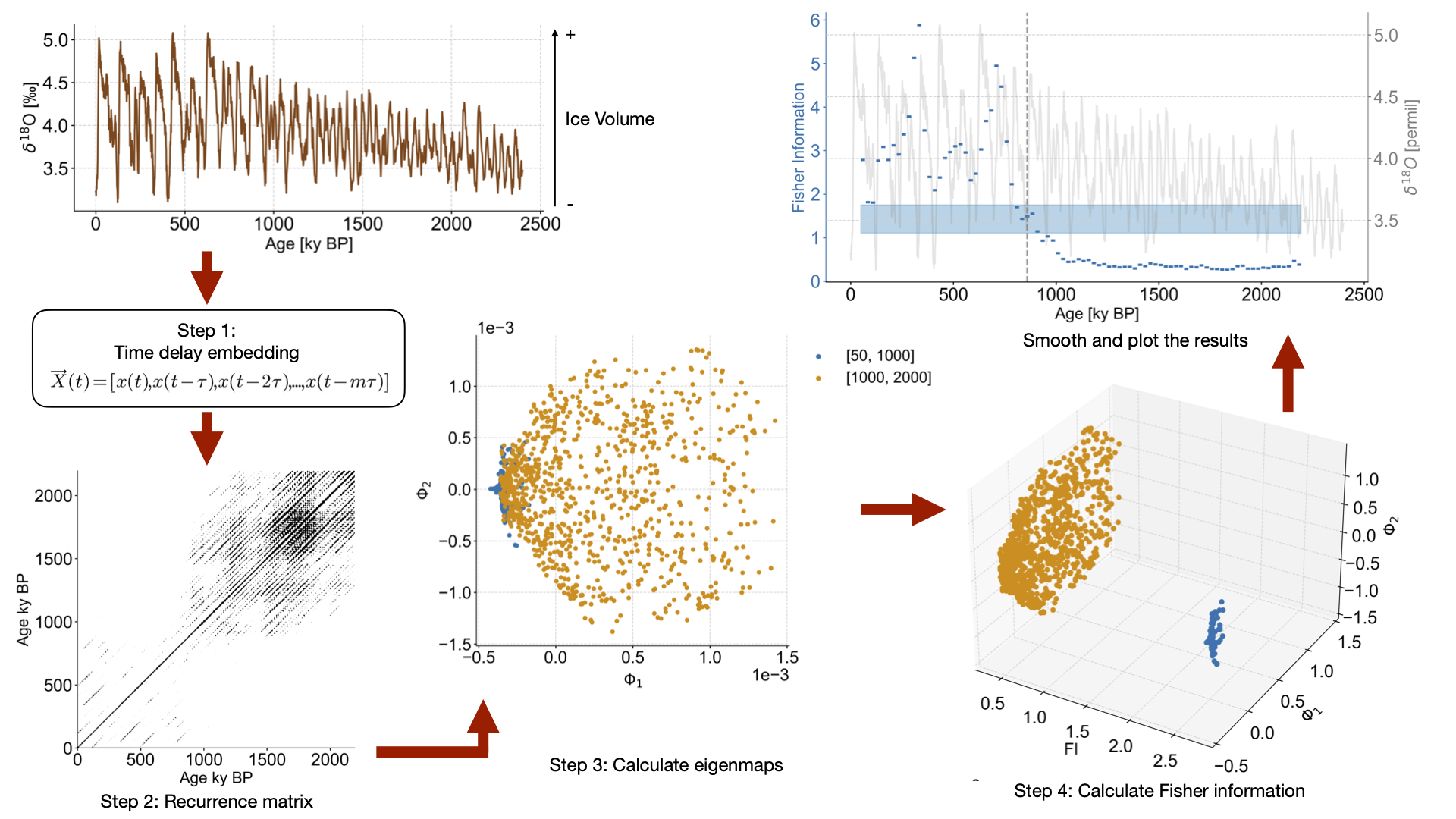
LERM applied to ODP cores#
This notebook recreates Figure 2 from the original publication, applying LERM to ODP cores over the Mid-Pleistocene-Transition (MPT).
import pyleoclim as pyleo
import matplotlib.pyplot as plt
from matplotlib import gridspec
import matplotlib.transforms as transforms
import matplotlib.patches as mpatches
import seaborn as sns
import numpy as np
import ammonyte as amt
from tqdm import tqdm
/Users/alexjames/miniconda3/envs/ammonyte/lib/python3.10/site-packages/pandas/core/arrays/masked.py:60: UserWarning: Pandas requires version '1.3.6' or newer of 'bottleneck' (version '1.3.5' currently installed).
from pandas.core import (
#We suppress warnings for these notebooks for presentation purposes. Best practice is to not do this though.
import warnings
warnings.filterwarnings('ignore')
#Defining group lists for easy loading
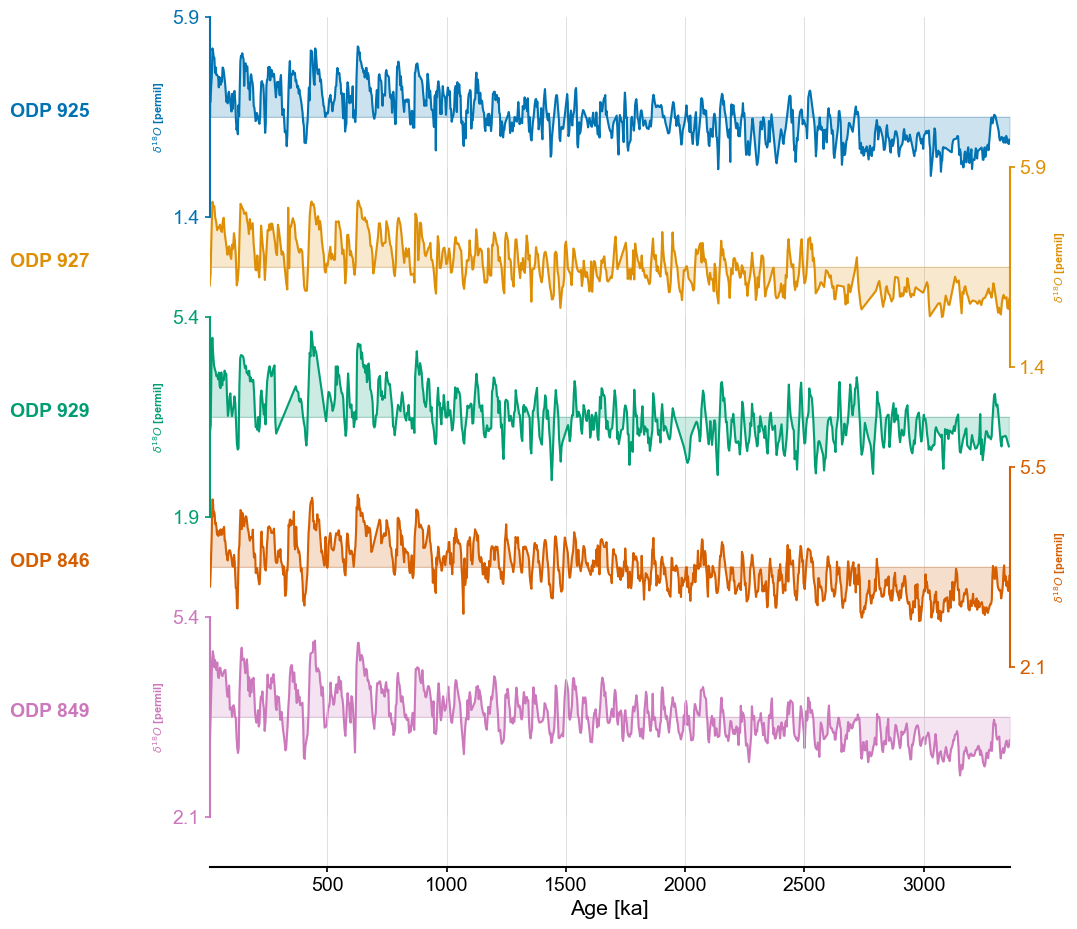
group_names = ['ODP 925','ODP 927','ODP 929','ODP 846','ODP 849']
Loading time series from LR04 stack data. The webpage with the data can be found here.
series_list = []
color_list = sns.color_palette('colorblind')
for name in group_names:
with open('../data/LR04cores_spec_corr/'+name[-3:]+'_LR04age.txt','rb') as handle:
lines = handle.readlines()
time = []
d18O = []
for x in lines:
line_time = float(format(float(x.decode().split()[1]),'10f'))
line_d18O = float(format(float(x.decode().split()[2]),'10f'))
#There is a discontinuity in 927 around 4000 ka, we'll just exclude it
if line_time <= 4000:
time.append(line_time)
d18O.append(line_d18O)
series = pyleo.Series(value=d18O,
time=time,
label=name,
time_name='Yr',
time_unit='ka',
value_name=r'$\delta^{18}O$',
value_unit='permil')
series_list.append(series)
max_time = min([max(series.time) for series in series_list])
min_time = max([min(series.time) for series in series_list])
ms = pyleo.MultipleSeries([series.slice((min_time,max_time)).interp() for series in series_list])
fig,ax = ms.stackplot(colors=color_list[:len(ms.series_list)],figsize=(8,10))
Time axis values sorted in ascending order
Time axis values sorted in ascending order
Time axis values sorted in ascending order
Time axis values sorted in ascending order
Time axis values sorted in ascending order
def detect_transitions(series,transition_interval=None):
'''Function to detect transitions across a confidence interval
Parameters
----------
series : pyleo.Series, amt.Series
Series to detect transitions upon
transition_interval : list,tuple
Upper and lower bound for the transition interval
Returns
-------
transitions : list
Timing of the transitions of the series across its confidence interval
'''
series_fine = series.interp(step=1)
if transition_interval is None:
upper, lower = amt.utils.sampling.confidence_interval(series)
else:
upper, lower = transition_interval
above_thresh = np.where(series_fine.value > upper,1,0)
below_thresh = np.where(series_fine.value < lower,1,0)
transition_above = np.diff(above_thresh)
transition_below = np.diff(below_thresh)
upper_trans = series_fine.time[1:][np.diff(above_thresh) != 0]
lower_trans = series_fine.time[1:][np.diff(below_thresh) != 0]
full_trans = np.zeros(len(transition_above))
last_above = 0
last_below = 0
for i in range(len(transition_above)):
above = transition_above[i]
below = transition_below[i]
if above != 0:
if last_below+above == 0:
loc = int((i+below_pointer)/2)
full_trans[loc] = 1
last_below=0
last_above = above
above_pointer = i
if below != 0:
if last_above + below == 0:
loc = int((i+above_pointer)/2)
full_trans[loc] = 1
last_above=0
last_below = below
below_pointer = i
transitions = series_fine.time[1:][full_trans != 0]
return transitions
Carrying out LERM analysis
lp_rm = {}
lp_fi = {}
m = 13 # Embedding dimension
for idx,series in enumerate(ms.common_time().series_list):
series = series.convert_time_unit('Years').interp().detrend(method='savitzky-golay')
amt_series = amt.Series(
time=series.time,
value=series.value,
time_name = series.time_name,
value_name = series.value_name,
time_unit = series.time_unit,
value_unit = series.value_unit,
label = series.label,
clean_ts=False,
sort_ts=None
)
td = amt_series.embed(m=m) # Tau is selected according to first minimum of mutual information
print(f'{series.label} tau is : {td.tau}')
eps = td.find_epsilon(eps = 1,target_density=.05,tolerance=.01) # Find epsilon value that gives us recurrence density of 5% (1 is the starting value we use for epsilon as a first guess)
rm = eps['Output']
lp_series = rm.laplacian_eigenmaps(w_size = 50,w_incre = 5) # Window size = 50, window increment = 5
lp_series = lp_series.convert_time_unit('ka')
lp_fi[series.label] = lp_series
ODP 925 tau is : 6
Initial density is 0.0232
Initial density is not within the tolerance window, searching...
Epsilon: 1.0000, Density: 0.0232
Epsilon: 1.1340, Density: 0.0537
Epsilon: 1.1340, Density: 0.0537.
ODP 927 tau is : 4
Initial density is 0.0365
Initial density is not within the tolerance window, searching...
Epsilon: 1.0000, Density: 0.0365
Epsilon: 1.0673, Density: 0.0533
Epsilon: 1.0673, Density: 0.0533.
ODP 929 tau is : 4
Initial density is 0.0410
Initial density is within the tolerance window!
ODP 846 tau is : 7
Initial density is 0.1141
Initial density is not within the tolerance window, searching...
Epsilon: 0.3590, Density: 0.0013
Epsilon: 0.8460, Density: 0.0386
Epsilon: 0.8460, Density: 0.0386
Epsilon: 0.9031, Density: 0.0603
Epsilon: 0.9031, Density: 0.0603
Epsilon: 0.8519, Density: 0.0405
Epsilon: 0.8519, Density: 0.0405.
ODP 849 tau is : 5
Initial density is 0.1123
Initial density is not within the tolerance window, searching...
Epsilon: 0.3768, Density: 0.0015
Epsilon: 0.8615, Density: 0.0415
Epsilon: 0.8615, Density: 0.0415.
SMALL_SIZE = 16
MEDIUM_SIZE = 20
BIGGER_SIZE = 25
plt.rc('font', size=SMALL_SIZE) # controls default text sizes
plt.rc('axes', titlesize=SMALL_SIZE) # fontsize of the axes title
plt.rc('axes', labelsize=MEDIUM_SIZE) # fontsize of the x and y labels
plt.rc('xtick', labelsize=MEDIUM_SIZE) # fontsize of the tick labels
plt.rc('ytick', labelsize=MEDIUM_SIZE) # fontsize of the tick labels
plt.rc('legend', fontsize=SMALL_SIZE) # legend fontsize
plt.rc('figure', titlesize=BIGGER_SIZE) # fontsize of the figure title
fig,axes = plt.subplots(nrows=len(group_names),ncols=1,sharex=True,figsize=(16,10))
transition_timing = []
for idx,site in enumerate(group_names):
ts = lp_fi[site]
ts.label = lp_series.label
ts.value_name = 'FI'
ts.value_unit = None
ts.time_name = 'Yr'
ts.time_unit = 'ka'
ax = axes[idx]
ts_smooth = amt.utils.fisher.smooth_series(series=ts,block_size=3) #Using a block size of 3 for smoothing the Fisher information
upper, lower = amt.utils.sampling.confidence_interval(series=ts,upper=95,lower=5,w=50,n_samples=10000) #Calculating the bounds for our confidence interval using default values
transitions=detect_transitions(ts_smooth,transition_interval=(upper,lower))
transition_timing.append(transitions[0])
ts.confidence_smooth_plot(
ax=ax,
background_series = ms.series_list[idx],
transition_interval=(upper,lower),
block_size=3,
color=color_list[idx],
figsize=(12,6),
legend=True,
lgd_kwargs={'loc':'upper left'},
hline_kwargs={'label':None},
background_kwargs={'ylabel':'$\delta^{18}O$ [permil]','legend':False,'linewidth':.8,'color':'grey','alpha':.8})
ax.axvline(transition_timing[idx],color='grey',linestyle='dashed',alpha=.5)
trans = transforms.blended_transform_factory(ax.transAxes, ax.transData)
ax.text(x=-.08, y = 2.5, s = site, horizontalalignment='right', transform=trans, color=color_list[idx], weight='bold',fontsize=20)
ax.spines['left'].set_visible(True)
ax.spines['right'].set_visible(False)
ax.yaxis.set_label_position('left')
ax.yaxis.tick_left()
ax.get_legend().remove()
ax.set_title(None)
ax.grid(visible=False,axis='y')
if idx != len(group_names)-1:
ax.set_xlabel(None)
ax.spines[['bottom']].set_visible(False)
ax.tick_params(bottom=False)
ax.xaxis.label.set_fontsize(25)
ax.yaxis.label.set_fontsize(25)
ax.set_yticks(ticks=np.array([0,5]))
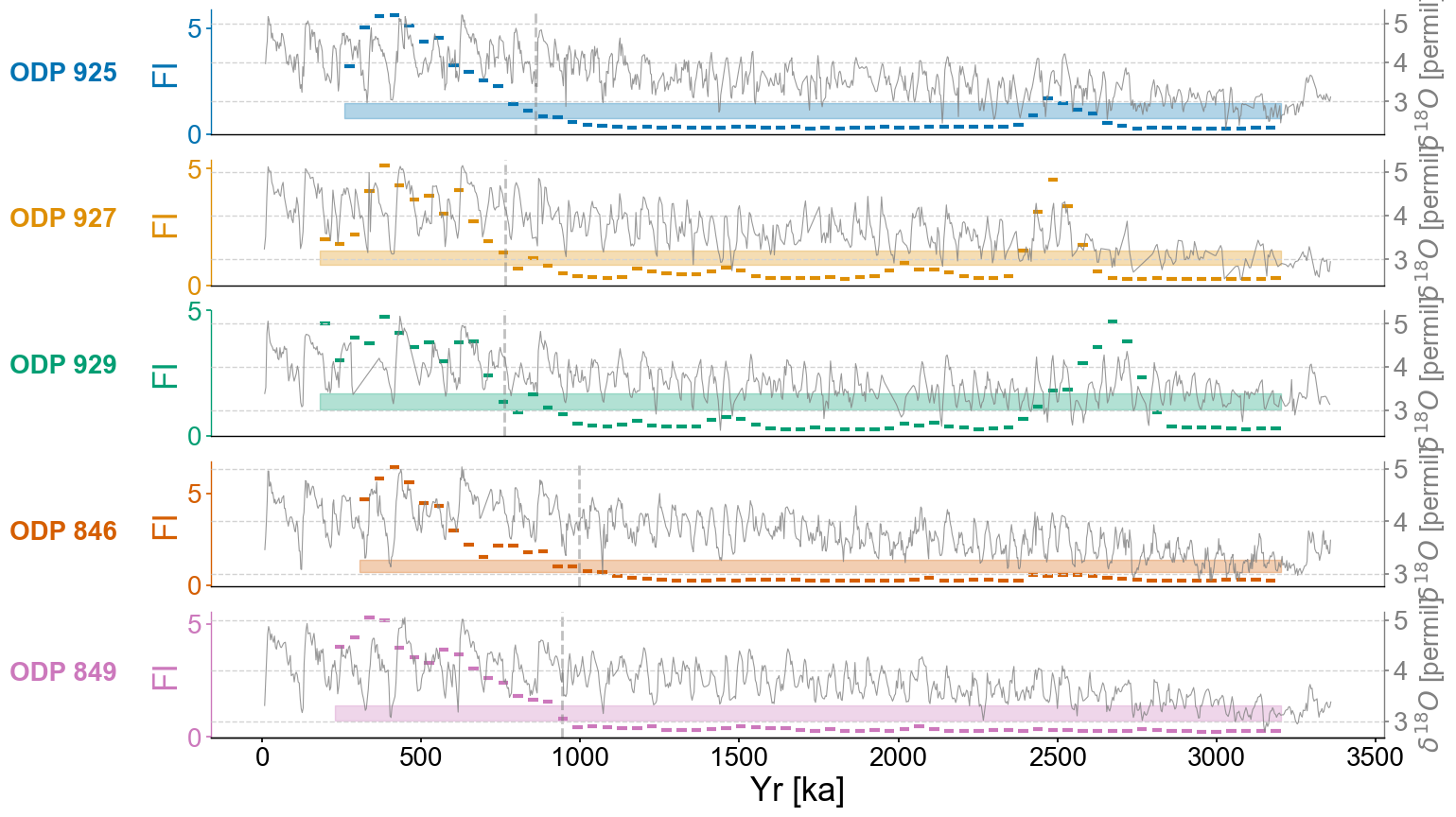
Checking the stats of the transition timings:
np.mean(transition_timing)
864.5573936631005
np.std(transition_timing)
94.32874897447839