Sensitivity to resolution#
This notebook recreates Figure 4 from the original publication, testing LERM’s sensitivity to resolution changes.
import pickle
import random
import pyleoclim as pyleo
import matplotlib.pyplot as plt
import matplotlib.patches as mpatches
import seaborn as sns
import numpy as np
import ammonyte as amt
/Users/alexjames/miniconda3/envs/ammonyte/lib/python3.10/site-packages/pandas/core/arrays/masked.py:60: UserWarning: Pandas requires version '1.3.6' or newer of 'bottleneck' (version '1.3.5' currently installed).
from pandas.core import (
#We suppress warnings for these notebooks for presentation purposes. Best practice is to not do this though.
import warnings
warnings.filterwarnings('ignore')
Top Panel#
#Defining group lists for easy loading
group_names = ['ODP 925']
series_list = []
color_list = sns.color_palette('colorblind')
for name in group_names:
with open('../data/LR04cores_spec_corr/'+name[-3:]+'_LR04age.txt','rb') as handle:
lines = handle.readlines()
time = []
d18O = []
for x in lines:
line_time = float(format(float(x.decode().split()[1]),'10f'))
line_d18O = float(format(float(x.decode().split()[2]),'10f'))
#There is a discontinuity in 927 around 4000 ka, we'll just exclude it
if line_time <= 4000:
time.append(line_time)
d18O.append(line_d18O)
series = pyleo.Series(value=d18O,
time=time,
label=name,
time_name='Yr',
time_unit='ka',
value_name=r'$\delta^{18}O$',
value_unit='permil')
series_list.append(series)
max_time = min([max(series.time) for series in series_list])
min_time = max([min(series.time) for series in series_list])
ms = pyleo.MultipleSeries([series.slice((min_time,max_time)).interp() for series in series_list])
fig,ax = ms.stackplot(colors=color_list[:len(ms.series_list)],figsize=(8,2*len(ms.series_list)))
Time axis values sorted in ascending order
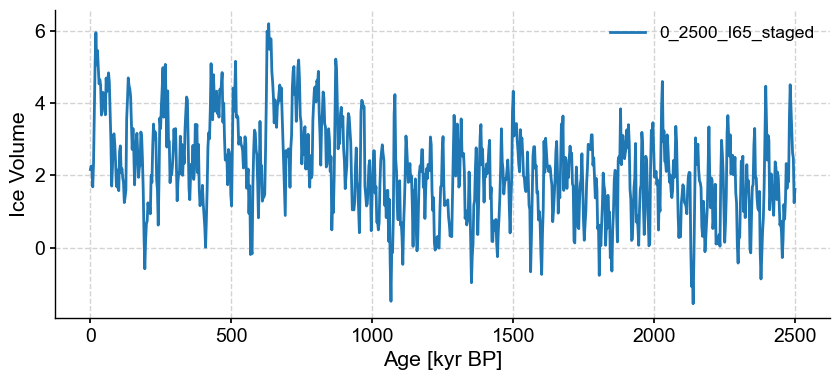
with open('../data/0_2500_I65_staged.pkl','rb') as handle:
initial_series = pickle.load(handle)
odp_series = ms.series_list[0]
binned_series = initial_series.bin(time_axis=odp_series.time) # Binning synthetic data onto the time axis of ODP 925
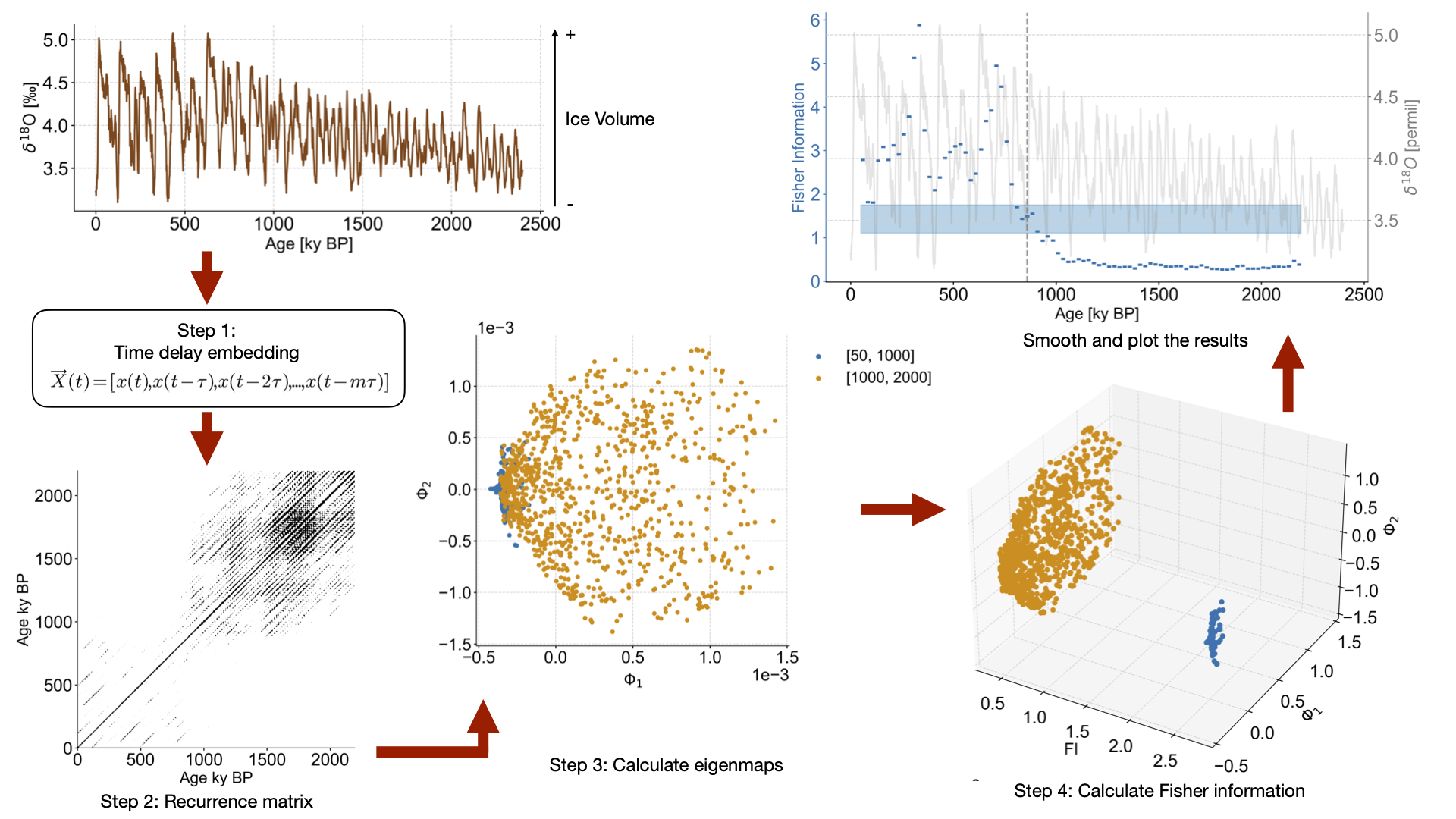
def noise_test(series,sn_ratio=1,m=13,tau=None,w_size=50,w_incre=5,find_eps_kwargs=None):
"""Run a noise test
Parameters
----------
series : pyleo.Series
Series on which to apply noise test
sn_ratio : float
Signal to noise ratio to use.
Note that signal to noise is defined as the standard deviation of the series
divided by the standard deviation of the noise
m : int
Embedding dimension
tau : int
Time delay embedding
w_size : int
Window size
w_incre : int
Window increment
find_eps_kwargs : dict
Dictionary of arguments for amt.TimeEmbeddedSeries.find_epsilon"""
find_eps_kwargs = {} if find_eps_kwargs is None else find_eps_kwargs.copy()
if 'eps' not in find_eps_kwargs:
find_eps_kwargs['eps'] = 1
noise_time,noise_value = pyleo.utils.gen_ts(model='ar1',t=series.time,scale=series.stats()['std']/sn_ratio) # Create AR1 series with appropriate standard deviation
noise = pyleo.Series(noise_time,noise_value,label='Noise Series',sort_ts=None) # Create noise series
real_sn_ratio = series.stats()['std']/noise.stats()['std'] # Calculate actual S/N ratio as sanity check
noisy_series = series.copy()
noisy_series.value += noise.value
amt_series = amt.Series(
time=noisy_series.time,
value=noisy_series.value,
time_name = noisy_series.time_name,
value_name = noisy_series.value_name,
time_unit = noisy_series.time_unit,
value_unit = noisy_series.value_unit,
label = noisy_series.label,
sort_ts=None
)
amt_series.convert_time_unit('Years')
td = amt_series.embed(m)
eps = td.find_epsilon(**find_eps_kwargs)
rm = eps['Output']
lp_series = rm.laplacian_eigenmaps(w_size=w_size,w_incre=w_incre)
lp_series.convert_time_unit('ka')
return lp_series, noisy_series, real_sn_ratio
sn_ratio = 2
find_eps_kwargs = {'eps':5}
# Here we use the noise_test function to carry out tests on the effect of the inclusion of noise.
# This function uses a default embedding dimension of 13, picks tau according to the first minimum of mutual information, and uses a default window size of 50 and window increment of 5
lp_series_2,noisy_series_2,real_sn_ratio_2 = noise_test(binned_series,
sn_ratio=sn_ratio,
find_eps_kwargs=find_eps_kwargs)
NaNs have been detected and dropped.
Initial density is 0.1851
Initial density is not within the tolerance window, searching...
Epsilon: 3.6487, Density: 0.0224
Epsilon: 3.9249, Density: 0.0376
Epsilon: 4.0493, Density: 0.0468
Epsilon: 4.0493, Density: 0.0468.
noisy_series_2.plot()
(<Figure size 1000x400 with 1 Axes>,
<Axes: xlabel='Age [kyr BP]', ylabel='Ice Volume'>)
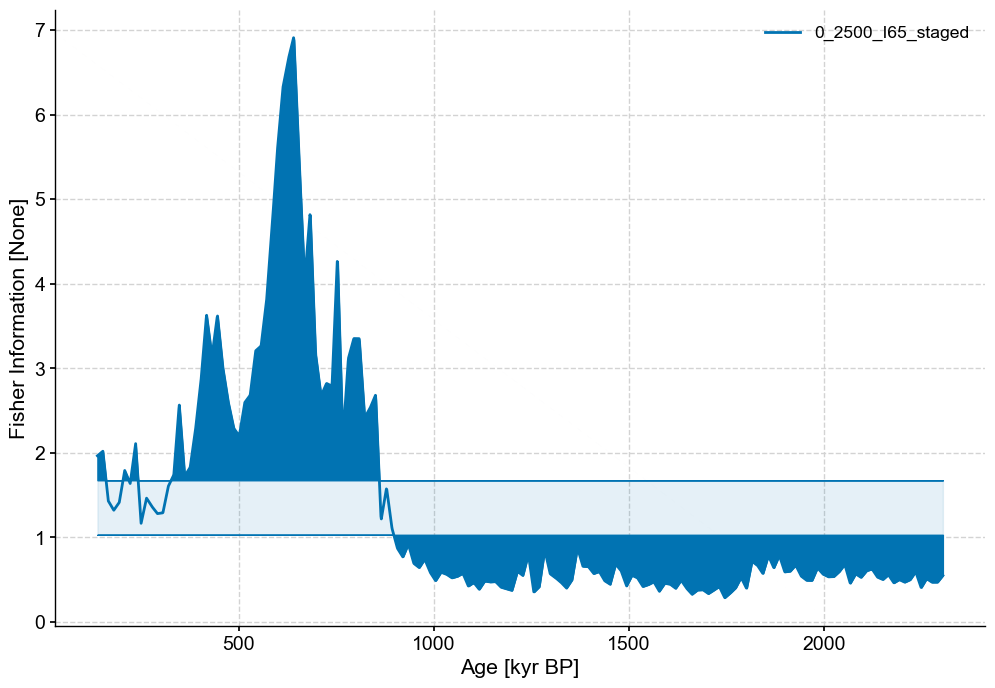
lp_series_2.confidence_fill_plot()
(<Figure size 1200x800 with 1 Axes>,
<Axes: xlabel='Age [kyr BP]', ylabel='Fisher Information [None]'>)
Middle panel#
with open('../data/0_2500_I65_stable.pkl','rb') as handle:
initial_series = pickle.load(handle)
stable_series = initial_series.bin(bin_size=4)
stable_series.label = 'Stable Series'
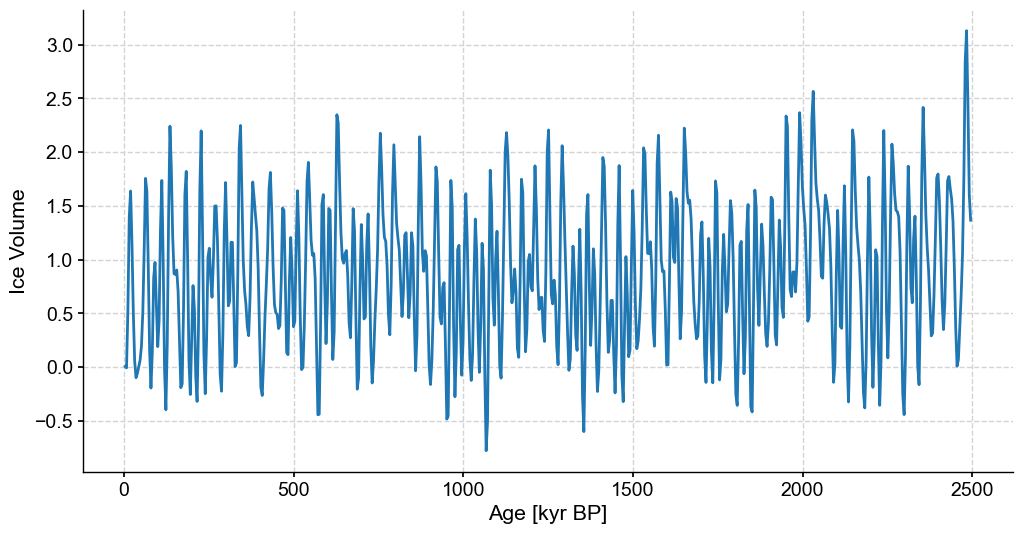
fig,ax = stable_series.plot(figsize=(12,6),legend=False)
series = stable_series
m=12 # Embedding dimension
amt_series = amt.Series(
time=series.time,
value=series.value,
time_name = series.time_name,
value_name = series.value_name,
time_unit = series.time_unit,
value_unit = series.value_unit,
label = series.label,
clean_ts=False
).convert_time_unit('Years')
td = amt_series.embed(m) # Pick tau according to first minimum of mutual info
eps = td.find_epsilon(1,.05,.01,parallelize=False) # Find epsilon that create recurrence matrix with density of 5% plus or minus 1%. Initial guess at epsilon is 1.
rm = eps['Output']
lp_stable = rm.laplacian_eigenmaps(50,5).convert_time_unit('ka') # Window size and window increment of 50 and 5 respectively
Time axis values sorted in ascending order
Initial density is 0.0022
Initial density is not within the tolerance window, searching...
Epsilon: 1.4781, Density: 0.0099
Epsilon: 1.8789, Density: 0.0401
Epsilon: 1.8789, Density: 0.0401.
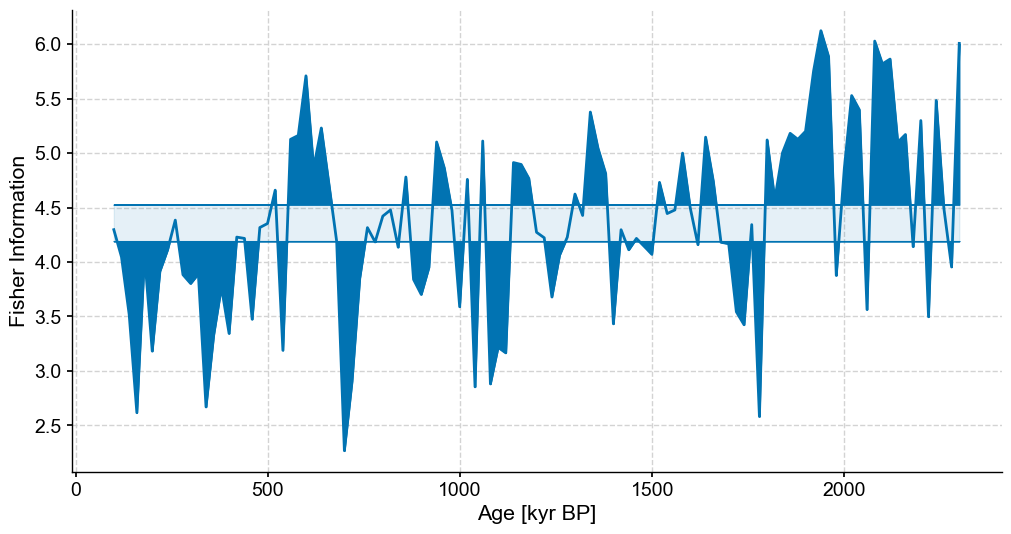
fig,ax = plt.subplots(figsize=(12,6))
lp_stable.confidence_fill_plot(ax=ax,legend=False)
ax.set_ylabel('Fisher Information')
ax.set_xlabel('Age [kyr BP]')
Text(0.5, 0, 'Age [kyr BP]')
Bottom panel#
def remove_values(series,percent,seed=42,subset=None):
'''Function to remove values at random
Note that this currently assumes units of Years BP
Parameters
----------
series : pyleoclim.Series
Series to remove values from
percent : float
Percentage of values to remove. Should be between 0 and 100
seed : int
Random seed to use
subset : list
[Start,stop] time indices for section of series you wish to be randomly coarsened.
If not passed, the whole series will be coarsened
Returns
-------
sparse_series : pyleoclim.Series
'''
random.seed(seed)
sparse_series = series.copy()
if subset:
sparse_slice = series.slice(subset)
sparse_index = np.where((series.time >= sparse_slice.time[0]) & (series.time <= sparse_slice.time[-1]))[0]
num_drop = int(len(sparse_slice.time) * (percent/100))
index_drop = sorted(random.sample(range(sparse_index[0],sparse_index[-1]), num_drop))
time = np.delete(series.time,index_drop)
value = np.delete(series.value,index_drop)
sparse_series.time = time
sparse_series.value = value
else:
num_drop = int(len(series.time) * (percent/100))
index_drop = sorted(random.sample(range(len(series.time)), num_drop))
time = np.delete(series.time,index_drop)
value = np.delete(series.value,index_drop)
sparse_series.time = time
sparse_series.value = value
return sparse_series
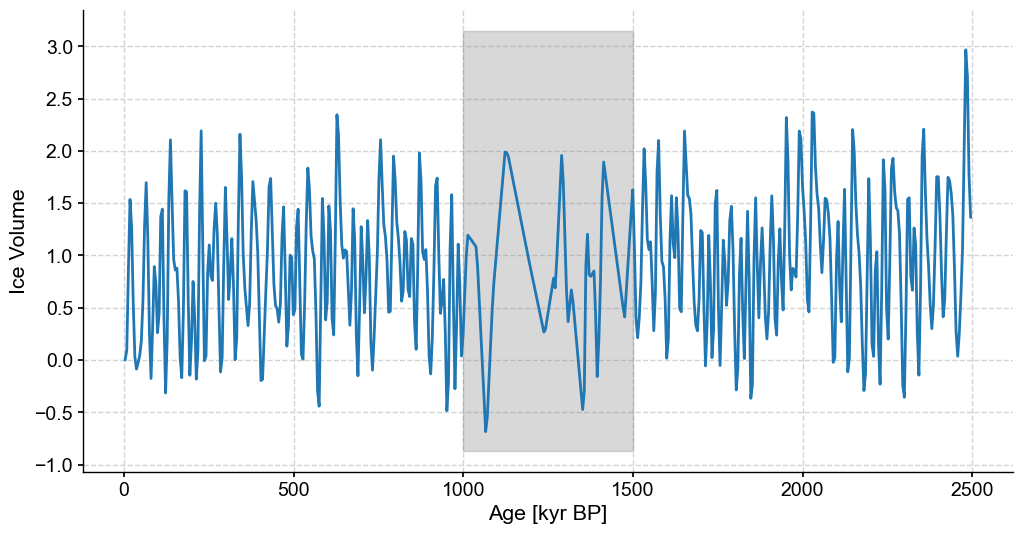
bounds = [1000,1500]
coarse_series = remove_values(series=stable_series,percent=80,subset=bounds).interp() #Artificially coarsening our stable synthetic data by removing 80% of values between specified bounds
fig,ax = coarse_series.plot(legend=False,figsize=(12,6))
ylim=ax.get_ylim()
ax.fill_betweenx(ylim,min(bounds),max(bounds),color='grey',alpha=.3,label='Coarse Section')
<matplotlib.collections.PolyCollection at 0x177d19780>
series = coarse_series.interp()
m=12
amt_series = amt.Series(
time=series.time,
value=series.value,
time_name = series.time_name,
value_name = series.value_name,
time_unit = series.time_unit,
value_unit = series.value_unit,
label = series.label,
clean_ts=False
).convert_time_unit('Years')
td = amt_series.embed(m=m)
eps = td.find_epsilon(eps=1,target_density=.05,tolerance=.01)
print(f'Tau is {td.tau}')
rm = eps['Output']
lp_coarse = rm.laplacian_eigenmaps(50,5).convert_time_unit('ka')
Time axis values sorted in ascending order
Initial density is 0.0029
Initial density is not within the tolerance window, searching...
Epsilon: 1.4708, Density: 0.0115
Epsilon: 1.8557, Density: 0.0470
Epsilon: 1.8557, Density: 0.0470.
Tau is 3
fig,ax = plt.subplots(figsize=(12,6))
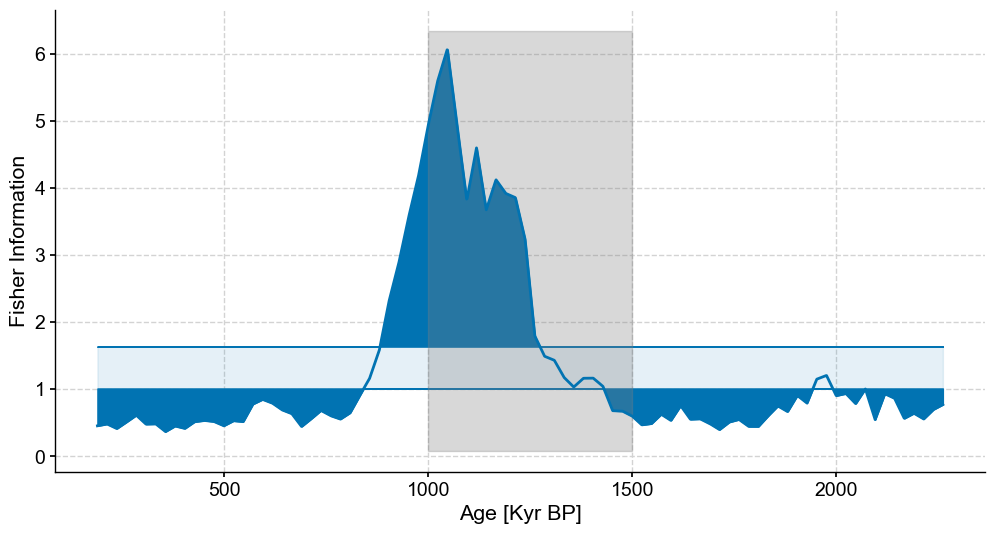
lp_coarse.confidence_fill_plot(ax=ax,legend=False)
ylim = ax.get_ylim()
ax.fill_betweenx(ylim,min(bounds),max(bounds),color='grey',alpha=.3,label='Coarse Section')
ax.set_ylabel('Fisher Information')
ax.set_xlabel('Age [Kyr BP]')
Text(0.5, 0, 'Age [Kyr BP]')
Full figure#
# Production figure
SMALL_SIZE = 26
MEDIUM_SIZE = 26
BIGGER_SIZE = 30
plt.rc('font', size=SMALL_SIZE) # controls default text sizes
plt.rc('axes', titlesize=MEDIUM_SIZE) # fontsize of the axes title
plt.rc('axes', labelsize=BIGGER_SIZE) # fontsize of the x and y labels
plt.rc('xtick', labelsize=MEDIUM_SIZE) # fontsize of the tick labels
plt.rc('ytick', labelsize=MEDIUM_SIZE) # fontsize of the tick labels
plt.rc('legend', fontsize=BIGGER_SIZE) # legend fontsize
plt.rc('figure', titlesize=BIGGER_SIZE) # fontsize of the figure title
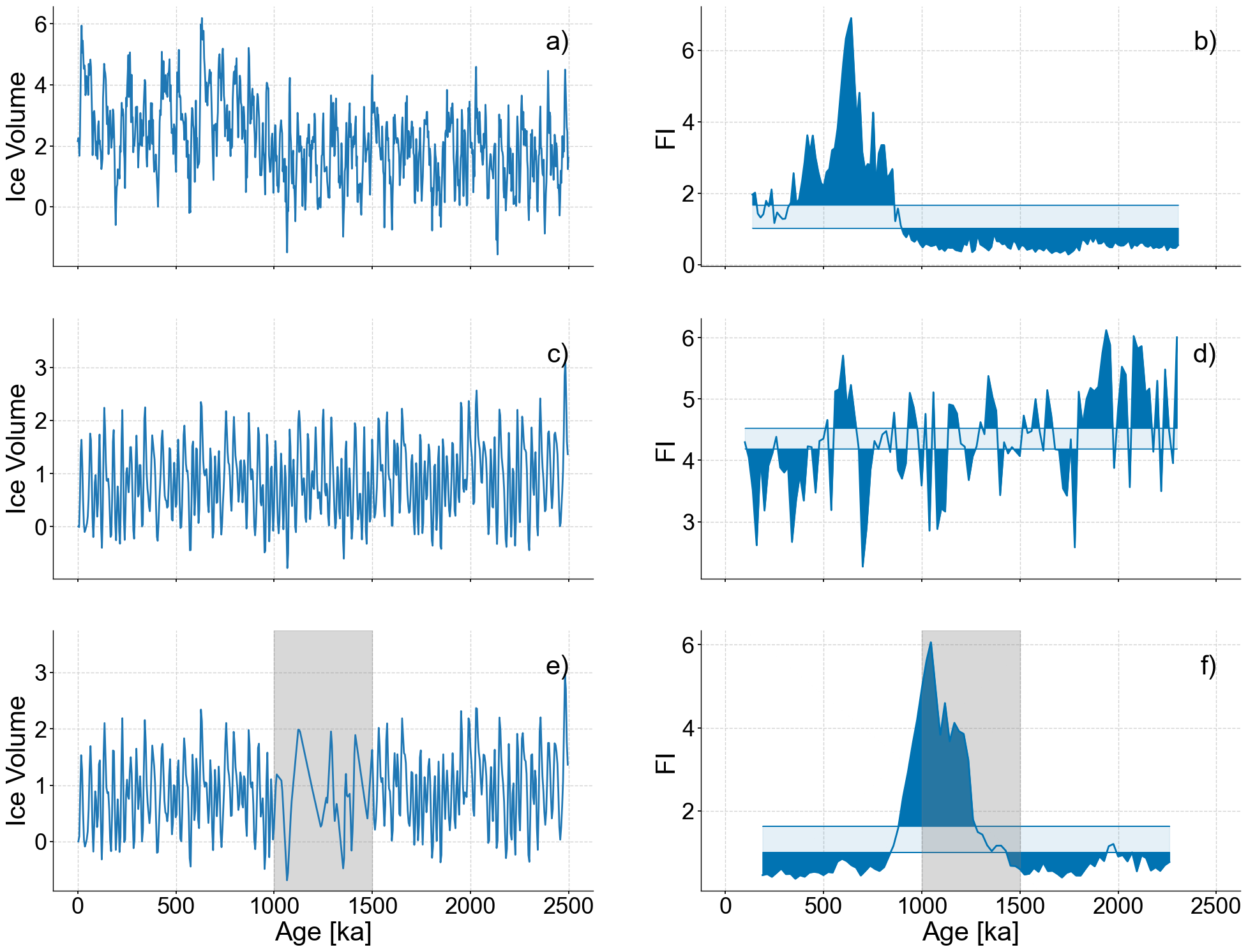
fig,axes = plt.subplots(nrows=3,ncols=2,sharex=True,figsize=(24,18))
noisy_series_2.plot(ax=axes[0,0],legend=None,xlabel='')
patch = mpatches.Patch(fc="w", fill=False, edgecolor='none', linewidth=0,label='a)')
axes[0,0].legend(handles=[patch],loc='upper right')
lp_series_2.confidence_fill_plot(ax=axes[0,1],legend=None,xlabel='',ylabel='FI')
patch = mpatches.Patch(fc="w", fill=False, edgecolor='none', linewidth=0,label='b)')
axes[0,1].legend(handles=[patch],loc='upper right')
stable_series.plot(ax=axes[1,0],legend=None,xlabel='')
ylim=list(axes[1,0].get_ylim())
ylim[1]+=.6
axes[1,0].set_ylim(ylim)
patch = mpatches.Patch(fc="w", fill=False, edgecolor='none', linewidth=0,label='c)')
axes[1,0].legend(handles=[patch],loc='upper right')
lp_stable.confidence_fill_plot(ax=axes[1,1],legend=None,xlabel='',ylabel='FI')
patch = mpatches.Patch(fc="w", fill=False, edgecolor='none', linewidth=0,label='d)')
axes[1,1].legend(handles=[patch],loc='upper right')
coarse_series.plot(ax=axes[2,0],legend=None,xlabel='Age [ka]')
ylim=list(axes[2,0].get_ylim())
ylim[1]+=.6
axes[2,0].fill_betweenx(ylim,min(bounds),max(bounds),color='grey',alpha=.3,label='Coarse Section')
axes[2,0].set_ylim(ylim)
patch = mpatches.Patch(fc="w", fill=False, edgecolor='none', linewidth=0,label='e)')
axes[2,0].legend(handles=[patch],loc='upper right')
lp_coarse.confidence_fill_plot(ax=axes[2,1],legend=None,ylabel='FI')
ylim=axes[2,1].get_ylim()
axes[2,1].fill_betweenx(ylim,min(bounds),max(bounds),color='grey',alpha=.3,label='Coarse Section')
axes[2,1].set_ylim(ylim)
patch = mpatches.Patch(fc="w", fill=False, edgecolor='none', linewidth=0,label='f)')
axes[2,1].legend(handles=[patch],loc='upper right')
<matplotlib.legend.Legend at 0x283fb38b0>